The determination of the total number of bacterial colonies is used to determine the degree of bacterial contamination of food and its hygienic quality. It reflects whether the food meets the hygienic requirements during the production and processing process, so as to make appropriate hygienic evaluation of the tested samples. The total number of bacterial colonies indicates the quality of food hygiene.
1. The deficiencies of GB4789.2 traditional plate counting agar method:
- The operation steps are cumbersome, and the preparation work takes a lot of time;
- Poor control of the pouring temperature of the medium can easily cause bacterial thermal damage;
- It is difficult to distinguish between colonies and sample impurities, and it is difficult to interpret;
Some samples were colonized and could not be counted.
Therefore, in this revision of the standard, the test piece is written into the national standard to meet the user’s requirements for an automated, simple, accurate and environmentally friendly method.
After nearly 10 years of development and optimization, the Meizheng total Aerobic Count Plate has passed the certification of AOAC and domestic authoritative institutions. The performance of plate count agar method consistency and so on.
2. Inclusive experimental results of the total Aerobic Count Plate method
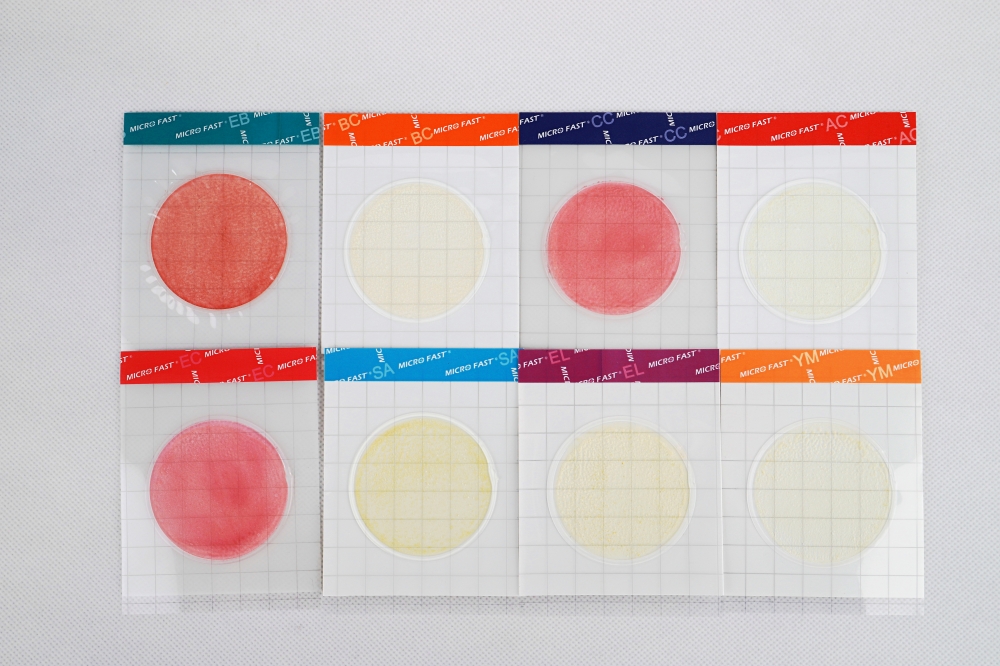
The results of each experimental strain showed that, except for Hansenula arabinitol (CGMCC 2.887), all other strains grew characteristic red colonies. The reason is that Hansenula arabinitol (CGMCC 2.887) is a fungus, not the target bacteria of the MicroFast®AC colony count test piece, so it cannot grow on the MicroFast®Aerobic Count Plate. The results of the inclusive experiment showed that the Aerobic Count Plate method had good inclusiveness.
Denaturation resistance test results of Aerobic Count Plate method.
After the test results were converted to 10 logarithms, the coefficient of variation (CV) of the MicroFast®AC colony count plate at different experimental temperatures were 2.64% and 2.62%, respectively. The test results have good repeatbility, indicating that the MicroFast®Aerobic Count Plate has good resistance to denaturation.
3. Repeatability test results of the Aerobic Count Plate method
After the test results were converted to 10 logarithms, the coefficients of variation of the test results of the bacterial suspensions of different concentrations and the naturally contaminated food samples with different matrices were all between 0.16% and 3.76%.
For artificially contaminated food samples, the coefficient of variation of the total number of colonies test piece method is 0.78%~8.61%, among which the coefficient of variation of ham sausage low-concentration and medium-concentration addition samples, ice cream low-concentration addition samples and milk powder high-concentration addition samples are greater than 5%. were 6.67%, 5.85%, 5.01% and 8.61%, respectively.
Except for the final concentration of sausage sausage, the national standard method also had a large coefficient of variation for the other three samples, and the coefficient of variation CV was 7.93%, 9.73% and 6.77%, respectively.
Possible reasons are: the presence of bacteriostatic agents in ham sausage, and the influence of milk protein in dairy products on the results of the total number of colonies counted by the two methods.
4. Method consistency evaluation results of the total Aerobic Count Plate method
18 kinds of sample matrices were selected and divided into naturally contaminated food samples and artificially contaminated food samples. The total number of colonies of the prepared bacterial suspensions, naturally contaminated food samples and artificially contaminated food samples were analyzed by the total number of colonies test piece method and plate counting agar method. The results showed that the difference between the two methods was not statistically significant (P>0.05), indicating that in different samples, the results of the two methods for total number of colonies were consistent.
View all Meizheng MicroFast® count plates.